In the three months or so that I’ve been working on this blog there has been some evolution in the methods I’m using. I though it would be worthwhile to revisit the first group I looked at to see if these changes in the methods affect my results. There have also been some additional sequences released since I started…
These methods are summarised here:
- Blast reference sequence against updated nt database (E-value cutoff 1e-180, max target sequences 3000): 310 results.
- Extract hit accession numbers from results and use blastdbcmd to acquire sequences from nt database.
- Use a bioperl script to duplicate redundant sequences: 521 sequences
- Use Eutils to download Genbank formatted sequences.
- Parse Genbank files to extract information on taxonomy, host and publication source for each sequence.
- Check that all sequences from included publications were extracted and add any missing sequences: 48 additional sequences (this was done manually)
- For studies that submitted a single representative sequence for each unique haplotype found, add duplicate entries for each specimen with that haplotype: 731 sequences total.
- Use uclust to group identical sequences and MAFFT to align non-redundant sequences.
- Use a bioperl script to identify alignment positions corresponding to gaps in a reference sequence, then use trimal to remove these from the alignment.
- Use phyml to infer a phylogeny and a perl script to map host information for each specimen onto it.
- Colour code tree by host family and view with FigTree.
Details of the commands for each of these steps are here. Files created at each step are here. The results of this process look like this:
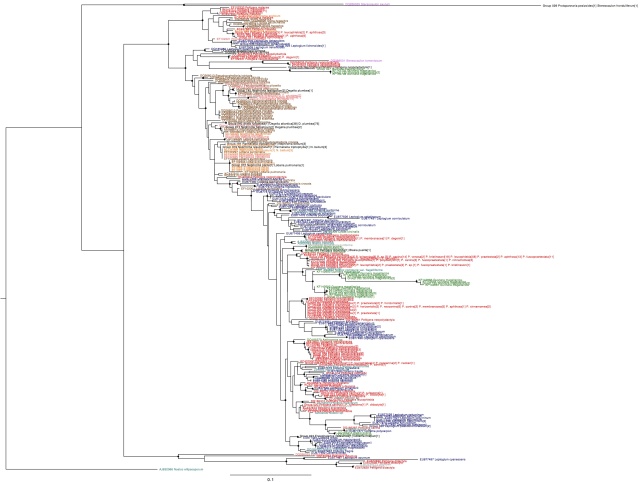
Nostoc rbcX phylogeny, coloured by host family (purple: Stereocaulaceae, brown: Lobariaceae, red: Peltigeraceae, blue: Collemetaceae, orange: Nephromataceae, pink: Pannariaceae, green: plant hosts, cyan: free-living). Names in black indicate genotypes found in more than one group. Circles on internal nodes indicate aLRT ≥0.9.
The total number of sequences included in the tree has increased from 547 to 731. These new sequences include photobionts of Stereocaulon exutum, S. fronduliferum, Protopannaria pezizoides, Fuscopannaria leucophaea, Leptogium azureum and L. cyanescens that form long branches at the base of the tree. The latter three branch with the early branching Peltigera didactlya sequences included in the previous analysis while the Stereocaulon and Protopannaria photobiont sequences form the sister group to all other photobiont sequences.
The topology of the tree is similar to the one produced previously, with a most symbionts of plants and lichens in the Peltigeraceae and Collemetaceae forming a large clade with Lobariaceae, Nephromataceae and Pannariaceae photobionts forming a paraphyletic grade at the base. Like the previous tree, there is also a clade of specialist photobionts that forms a sister group to this lineage. The major difference is that a group of rapidly evolving symbionts of Peltigera neopolydactyla, Massalongia carnosa, Stereocaulon tomentosum and the flowering plant Gunnera magellanica that were nested within the main Peltigeraceae/Collemetaceae/plant symbiont clade in the previous analysis now branch at the base of the main photobiont clade.
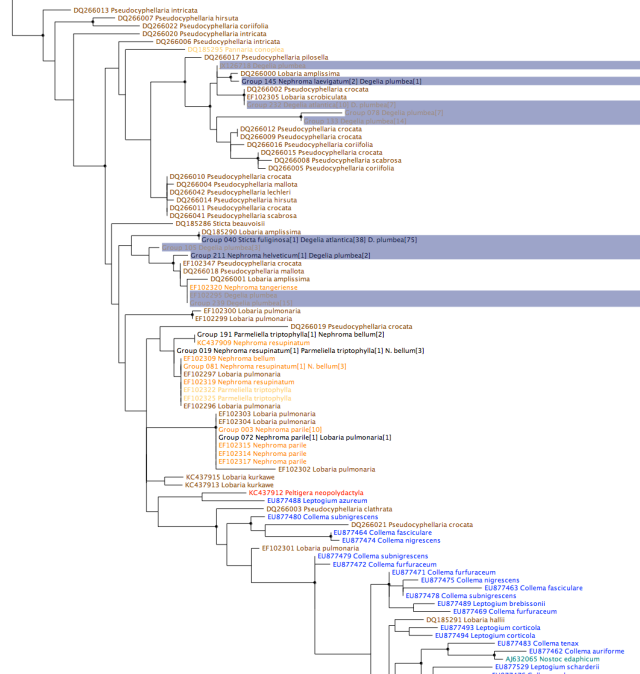
Many of the new sequences that have been added in this analysis are from this paper which compared the photobiont diversity of closely related sexual and asexual species. They found nine different photobiont haplotypes in the sexual species (Degelia plumbea) versus only two for the asexual species (Degelia atlantica). This can clearly be seen by zooming in on the basal Lobariaceae/Nephromataceae/Pannariaceae grade in the tree and highlighting Degelia photobionts:
D. atlantica is restricted to one photobiont genotype shared with D. plumbea, Lobaria scrobiculata and Pseudocyphellaria crocata photobionts and one shared with photobionts of D. plumbea, Lobaria amplissima and Sticta fuliginosa, while D. plumbea photobionts are much more diverse. This is a dramatic difference and confirms a hypothesis about the interaction of asexuality, photobiont specificity and narrow habitat requirements presented previously. However, the D. plumbea photobionts form two clusters of closely related genotypes, with one D. atlantica photobiont genotype in each cluster. This suggests that both species may have similar photobiont specificity determinants at a genetic level, but that codispersal has resulted in limited opportunities for switching to other closely related photobiont genotypes. I am sceptical that the ecological niche of D. atlantica is limited by the genetic diversity of its photobionts since photobionts from the two different clusters are much more likely to have different growth requirements than photobionts within clusters, but this is something that will ultimately have to be tested experimentally.
Heath OBrien (2013). Symbiotic Nostoc Revisited PhotobiontDiversity.wordpress.com : http://dx.doi.org/10.6084/m9.figshare.775384

Pingback: Another perspective on diversity of symbiotic cyanobacteria: 16S | PhotobiontDiversity
Pingback: Peltigera Canina | Find Me A Cure